Biology & Applied Physics, Ultrasound Imaging, Biosciences, Radiation Imaging, Magnetic Resonance Physics, Artificial Intelligence, Machine Learning, Computer Vision
According to the World Health Organization[i], 2.1 million world-wide are diagnosed with breast cancer every year. In the US alone, the number of cases diagnosed is estimated to be more than 250,000 with more than 40,000 deaths[ii]. The survival rate in the US and other more developed nations is improving however, thanks in part to early diagnosis and more effective treatments [iii],[iv].
Hear from two scientists at GE Research, Fiona Ginty, biosciences technology manager, and Cynthia Davis, a radiation physics principal scientist, on how their work is helping to advance breast cancer research and forge new boundaries in the cancer imaging space.
How has technology for breast cancer changed over the last 20 years?
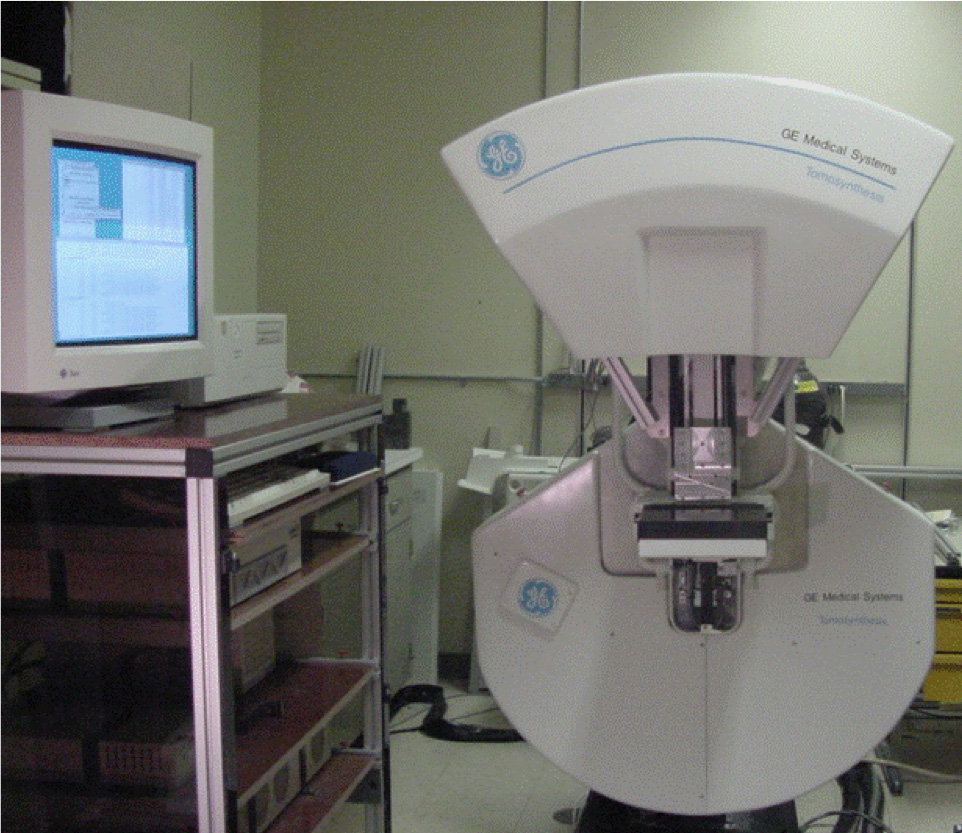
Cindy Davis: Breast imaging has seen tremendous changes in the past 20 years. We have gone from film-based technology to digital technology. I’m proud to say that when I joined GRC I was on the team to bring to market the first digital mammography system[v]. Digital mammography improved detection in younger women and women with dense breasts vs screen film in particular[vi]. Then we shifted our attention to tomosynthesis, a 3D breast imaging technology and GE installed the first tomosynthesis system at Massachusetts General Hospital (MGH)[vii]. Today tomosynthesis has become the standard of care for breast imaging.
Fiona Ginty: Similarly, in tumor biology and diagnostics, there have been many advances in the last 20 years. Testing for hormone receptors and Her2 has been a standard part of patient diagnostic workflow and treatments planning.
CD: Another area where GE has led is automated breast ultrasound (ABUS). With our collaboration with the University of Michigan we have developed ultrasound fusion systems that paved the way for our Invenia ABUS products[viii]. Our team here at GE Research has also worked on breast MRI and making exams shorter and improving the specificity[ix]. We have also worked on contrast enhanced mammography (and tomosynthesis) which should bring new clarity for diagnosis.
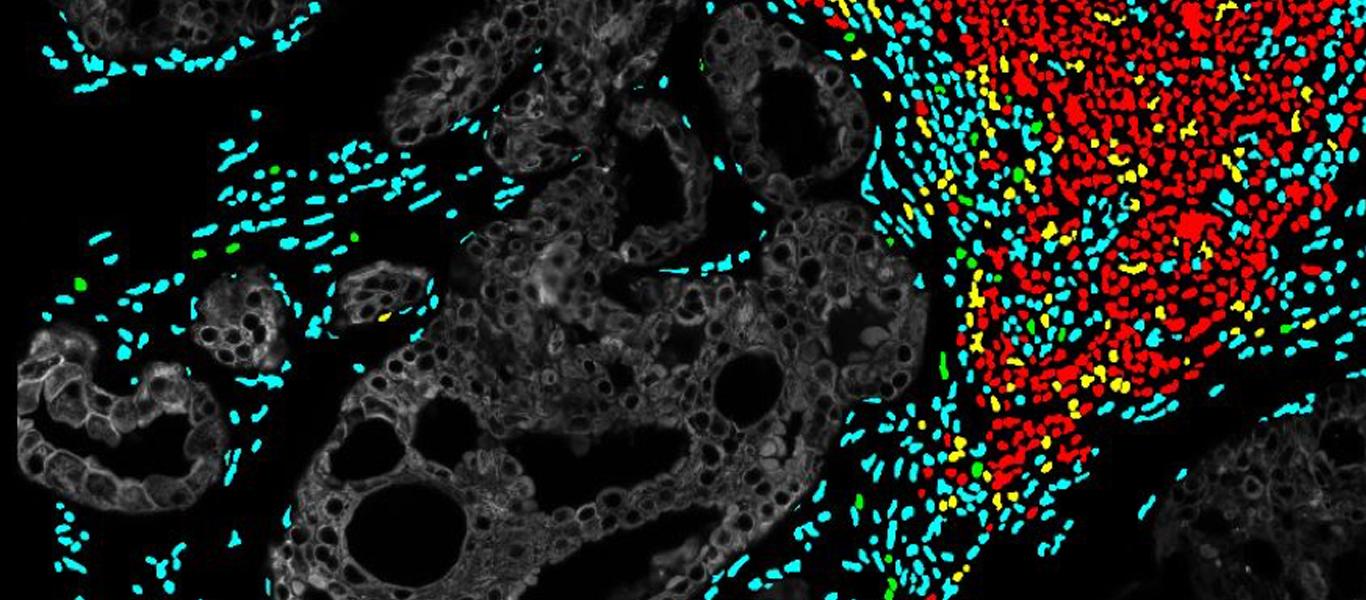
FG: Most recently the focus has been on genetic and cellular composition/heterogeneity of tumors and how they contribute to disease progression. At GE Research, together with Indiana University, we have been funded by the NIH (R01CA194600-04) to investigate the cellular composition of Ductal Carcinoma in situ (DCIS), using a novel analysis platform “Cell DIVE” developed by our Scientists and Engineers over 10 years and now commercially launched by GEHC.
Using this detailed cellular and spatial information, we can begin to understand the mechanisms of why some cancers are more aggressive than others, how the immune system responds in different populations and how tumor heterogeneity plays a role [6].
One of the issues facing breast cancer today is overtreatment of cancers. What is the current research to help determine which cancers we need to aggressively treat?
CD: With advances of screening we are finding cancers earlier and earlier, much before they can be felt. However, it is difficult to determine if a particular cancer will spread beyond the breast and hence become deadly. We know that certain cancers are more aggressive such as triple negative (Estrogen receptor, progesterone receptor negative and Her2 negative). There is a large body of research to identify imaging characteristics to predict the likelihood of progression and help direct treatment.
FG: Our cutting-edge research in tumor cell imaging and hyperplex in situ proteomic analysis, use of artificial intelligence to unearth hidden clues, quantification of data analytics will help delineate some previously unknown mechanisms and characteristics of aggressive disease. As part of our collaboration with Indiana University, we are also investigating differences in DCIS between Caucasian, Asian and African American women and the underlying mechanisms. For example, African American women have a higher prevalence of triple negative breast cancer, which is a more aggressive form of breast cancer. We are measuring the cellular composition of DCIS lesions as well as gene expression differences to better understand the characteristics of aggressive disease and potential drug targets.
Precision medicine is the next technology frontier for breast cancer research. What are some of the enabling technologies that GE is working on that will enable more precise management of breast health?
CD: One area for precision medicine is improving breast cancer screening with information of risk profile not just age. Women with BRCA or other genetic pre-disposition for breast cancer have long had MRI screening performed. Recently, breast density has been identified as an additional risk factor for breast cancer as well as decreased efficacy of mammography screening. Women with dense breasts in many states can have additional supplemental screening based upon this[x]. In the future as risks for breast cancer are found, screening can become more and more personalized to woman’s risk not just her age.
FG: Liquid biopsy is another new approach to monitor tumor changes post treatment. This involves the separation of a variety of soluble biomarkers (e.g. tumor cells, mutations or methylation status of cell free DNA (cfDNA), RNA and protein expression) from various bodily fluids. This provides a non-invasive way of monitoring changes that are specific to tumors and potentially provide an early indicator of treatment response or lack thereof. A common bodily fluid used for liquid biopsy is blood and the commonly used biomarkers are mutations and methylation status of cfDNA. The challenge is that the amount of cfDNA is very low and requires several tubes of blood to get sufficient material. In collaboration between GE Research and U. Pittsburgh, our team has applied a novel DNA amplification method and tested it in blood from patients with ER-positive metastatic breast cancer and found that all the major mutations are detected in a small amount of blood [7]. This work is still at the early stages, but shows the potential of DNA monitoring for patients.
[i] World Health Organization, “Breast Cancer”, https://www.who.int/cancer/prevention/diagnosis-screening/breast-cancer/en/ accessed Oct. 24, 2019.
[ii] American Cancer Society, “Breast Cancer facts and figures 2017-2018, https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/breast-cancer-facts-and-figures/breast-cancer-facts-and-figures-2017-2018.pdf accessed Oct. 24, 2019.
[iii] Van Schoor G, Moss SM, Otten JD, et al. Increasingly strong reduction in breast cancer mortality due to screening. Br J Cancer 104:910-914, 2011.
[iv] Marmot MG, Altman DG, Cameron DA, Dewar JA, Thompson SG, Wilcox M. The benefits and harms of breast cancer screening: an independent review. Br J Cancer. 2013;108(11):2205-2240.
[v] Vedantham S, Karellas A, Suryanarayanan S, Albagli D, Han S, Tkaczyk EJ, Landberg CE, Opsahl-Ong B, Granfors PR, Levis I, D'Orsi CJ, Hendrick RE. Full breast digital mammography with an amorphous silicon-based flat panel detector: physical characteristics of a clinical prototype. Med Phys. 2000 Mar;27(3):558-67.
[vi] Pisano ED, Gatsonis C, Hendrick E, Yaffe M, Baum JK, Acharyya S, Conant EF, Fajardo LL, Bassett L, D'Orsi C, Jong R, Rebner M, Digital Mammographic Imaging Screening Trial (DMIST) Investigators Group. Diagnostic performance of digital versus film mammography for breast-cancer screening. N Engl J Med. 2005 Oct 27; 353(17):1773-83.
[vii] Niklason LT, Christian BT, Niklason LE, Kopans DB, Castleberry DE, Opsahl-Ong BH, Landberg CE, Slanetz PJ, Giardino AA, Moore R, Albagli D, DeJule MC, Fitzgerald PF, Fobare DF, Giambattista BW, Kwasnick RF, Liu J, Lubowski SJ, Possin GE, Richotte JF, Wei CY, Wirth RF. Digital tomosynthesis in breast imaging. Radiology. 1997 Nov;205(2):399-406.
[viii] Kapur A, Carson PL, Eberhard J, Goodsitt MM, Thomenius K, Lokhandwalla M, Buckley D, Roubidoux MA, Helvie MA, Booi RC, LeCarpentier GL, Erkamp RQ, Chan HP, Fowlkes JB, Thomas J, Landberg C. Combination of digital mammography with semi-automated 3D breast ultrasound. Technol Cancer Res Treat. 2004 Aug;3(4):325-34.
[ix] Thakur, S. B., J. V. Horvat, et al. (2019). "Quantitative in vivo proton MR spectroscopic assessment of lipid metabolism: Value for breast cancer diagnosis and prognosis." J Magn Reson Imaging 50(1): 239-249.
[6] Gerdes, Michael J., Yesim Gökmen-Polar, Yunxia Sui, Alberto Santamaria Pang, Nicole LaPlante, Adrian L. Harris, Puay-Hoon Tan, Fiona Ginty, and Sunil S. Badve. "Single-cell heterogeneity in ductal carcinoma in situ of breast." Modern Pathology 31, no. 3 (2018): 406.
[7] Gyanchandani, Rekha, Erik Kvam, Ryan Heller, Erin Finehout, Nicholas Smith, Karthik Kota, John R. Nelson et al. "Whole genome amplification of cell-free DNA enables detection of circulating tumor DNA mutations from fingerstick capillary blood." Scientific reports 8, no. 1 (2018): 17313.
[x] https://www.cancer.gov/types/breast/breast-changes/dense-breasts
We're here to solve your toughest problems.